19A123
PROTEIN BIOMARKERS TO DIFFERENTIATE PSORIATIC ARTHRITIS FROM PSORIASIS
Author(s)
Conor Magee; Yitong Pu; Anna Kwasnik; Angela McArdle; Belinda Hernandez; Stephen Pennington; Oliver FitzGerald
Department(s)/Institutions
The Conway Institute, UCD; St. Vincent's University Hospital
Introduction
In the BIOmarkers of COMorbidities (BIOCOM) in Psoriasis (Pso) study we aim to identify clinical, genetic and protein biomarker features associated with the development of psoriatic arthritis (PsA), in patients with Pso. Pso usually precedes the development of PsA with an average interval of 10 years. Thus, patients with Pso are an ideal group in which to study the early events in the evolution to PsA.
Aims/Background
To use a targeted proteomics approach to identify serum proteins which can predict the development of PsA in patients with Pso. We initially sought to identify serum proteins capable of discriminating between patients with Pso only and patients with established PsA.
Method
30 patients with Pso and 30 patients with established PsA were selected from the BIOCOM-Pso
database. Serum samples from these patients were digested using a standard operating protocol (SOP). Once digested, a targeted proteomics approach using liquid chromatography – mass spectrometry (LC-MS) and a multiple reaction monitoring (MRM) assay called PAPRICA was used to measure candidate biomarker proteins. These 206 proteins (423 peptides) were previously identified as being potential biomarkers in a number of different inflammatory rheumatological conditions.
Results
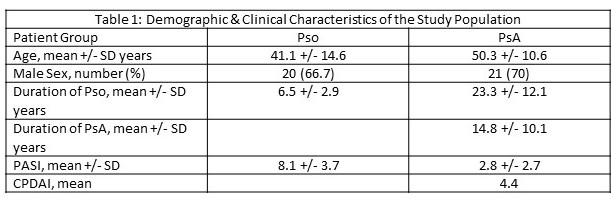
The demographics of the 2 patient groups are shown in Table 1.
The initial results revealed that the application of the PAPRICA method to the BIOCOM-Pso samples resulted in a dataset in which 275 of the 423 PAPRICA peptides could be reliably measured (CV Area < 20%; Signal to Noise ratio > 5; Library Dot Product > 0.8). Targeted proteomics data from the PsA and Pso patients was subjected to univariate and multivariate analysis. Univariate analysis revealed five peptides with a p value < 0.05, however none of these remained significant after Bonferroni correction for multiple comparisons. Multivariate analysis was unable to discriminate between PsA and Pso.
Conclusions
Analysis of the 206 biomarker proteins in the PAPRICA method, in patients with PsA and Pso, did not reveal peptides (proteins) that were statistically different between these two
groups. The next steps will include supplementing the PAPRICA method with additional biomarkers including proteins that may be identified in an unbiased proteome wide screen of PsA vs Pso serum samples.